Vector search has unlocked the door to another level of relevance and efficiency in information retrieval. In the past year, the number of vector search use cases has exploded, showing no signs of slowing down.
The capabilities of vector search are impressive, but it isn’t a perfect technology. In fact, without big domain-specific datasets to fine-tune models on, a traditional search still has some advantages.
We repeatedly see that vector search unlocks incredible and intelligent retrieval but struggles to adapt to new domains. Whereas traditional search can cope with new domains but is fundamentally limited to a set performance level.
Both approaches have pros and cons, but what if we merge them somehow to eliminate a few of those cons? Could we create a hybrid search with the heightened performance potential of vector search and the zero-shot adaptability of traditional search?
Today, we will learn how to take our search to a new level. Taking both vector and traditional search and merging them via Pinecone’s new hybrid search.
Out of Domain Datasets
Vector search or dense retrieval has been shown to significantly outperform traditional methods when the embedding models have been fine-tuned on the target domain. However, this changes when we try using these models for “out-of-domain” tasks.
That means if we have a large amount of data covering a specific domain like “Medical question-answering”, we can fine-tune an embedding model. With that embedding model, we can create dense vectors and get outstanding vector search performance.
The problem is if we don’t have data. In this scenario, a pretrained embedding model may perform better than traditional BM25, but it is unlikely. Giving us a best-case performance of BM25, an algorithm that we cannot fine-tune and cannot provide intelligent human-like retrieval.
If we want better performance, we’re left with two options; (1) annotate a large dataset to fine-tune the embedding model, or (2) use hybrid search.
Hybrid Search
Combining dense and sparse search takes work. In the past, engineering teams needed to run different solutions for dense and sparse search engines and another system to combine results in a meaningful way. Typically a dense vector index, sparse inverted index, and reranking step.
The Pinecone approach to hybrid search uses a single sparse-dense index. It enables search across any modality; text, audio, images, etc. Finally, the weighting of dense vs. sparse can be chosen via the alpha parameter, making it easy to adjust.
How does a hybrid search pipeline look?
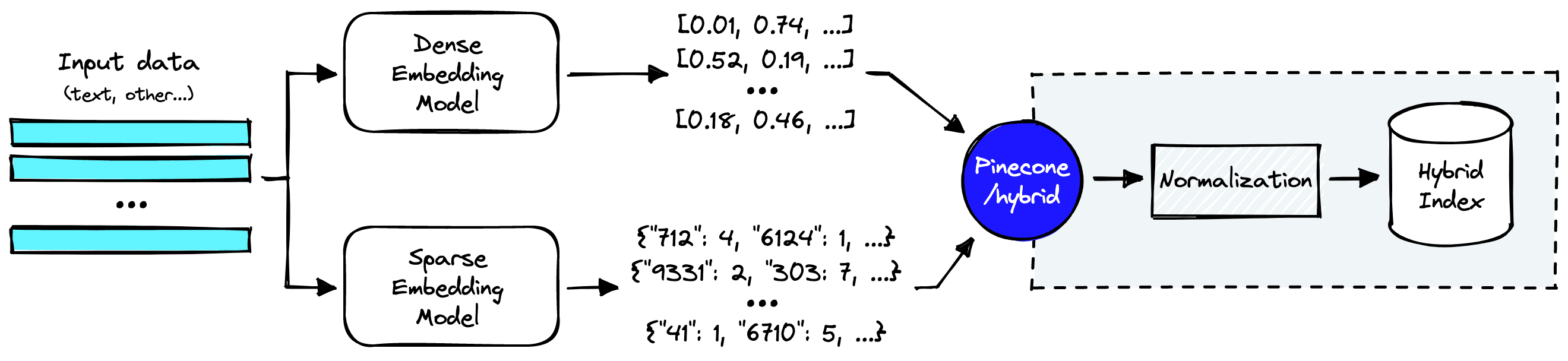
Everything within the dotted lines is handled by Pinecone’s hybrid index. But before we get there, we still need to create dense and sparse vector representations of our input data.
Let’s take a look at how we can do that.
Implementation of Hybrid Search
The first step in a hybrid search implementation is preparing a dataset. We will use the pubmed_qa dataset on Hugging Face Datasets. We download it like so:
from datasets import load_dataset # !pip install datasets
pubmed = load_dataset(
'pubmed_qa',
'pqa_labeled',
split='train'
)
pubmedDataset({ features: ['pubid', 'question', 'context', 'long_answer', 'final_decision'], num_rows: 1000 })
The context feature is what we will store in Pinecone. Each context record contains multiple contexts within a list. Many lack real significance alone, so we will join them to create larger contexts.
contexts = []
# loop through the context passages
for record in pubmed['context']:
# join context passages for each question and append to contexts list
contexts.append('\n'.join(record['contexts']))
# view some of the contexts
for context in contexts[:2]:
print(f"{context[:300]}...")Programmed cell death (PCD) is the regulated death of cells within an organism. The lace plant (Aponogeton madagascariensis) produces perforations in its leaves through PCD. The leaves of the plant consist of a latticework of longitudinal and transverse veins enclosing areoles. PCD occurs in the cel... Assessment of visual acuity depends on the optotypes used for measurement. The ability to recognize different optotypes differs even if their critical details appear under the same visual angle. Since optotypes are evaluated on individuals with good visual acuity and without eye disorders, differenc...
We can see the highly-technical language contained within each context. An out-of-the-box model will typically struggle with this domain-specific language, making this an ideal use-case for hybrid search.
Let’s move on to building the sparse and dense vectors.
Sparse Vectors
Several methods exist for building sparse vector embeddings, from the latest sparse embedding transformer models like SPLADE to rule-based tokenization logic.
We will stick with a more straightforward tokenization approach to keep things simple. Like the BERT tokenizer hosted by Hugging Face Transformers.
from transformers import BertTokenizerFast # !pip install transformers
# load bert tokenizer from huggingface
tokenizer = BertTokenizerFast.from_pretrained(
'bert-base-uncased'
)To tokenize a single context, we can do this:
# tokenize the context passage
inputs = tokenizer(
contexts[0], padding=True, truncation=True,
max_length=512
)
inputs.keys()dict_keys(['input_ids', 'token_type_ids', 'attention_mask'])
The output from this includes a few arrays that are all important when using transformer models. As we’re doing tokenization only, we need the input_ids.
input_ids = inputs['input_ids']
input_ids[101, 16984, 3526, 2331, 1006, 7473, 2094, ...]
These input IDs represent a unique word or sub-word token translated into integer ID values. This transformation is done using the BERT tokenizer’s rule-based tokenization logic.
Pinecone expects to receive sparse vectors in dictionary format. For example, the vector:
[0, 2, 9, 2, 5, 5]
Would become:
{ "0": 1, "2": 2, "5": 2, "9": 1 }
Each token is represented by a single key in the dictionary, and its frequency is counted by the respective key-value. We apply the same transformation to our input_ids like so:
from collections import Counter
# convert the input_ids list to a dictionary of key to frequency values
sparse_vec = dict(Counter(input_ids))
sparse_vec{101: 1, 16984: 1, 3526: 2, 2331: 2, 1006: 10, ... }
We can reformat all of this logic into two functions; build_dict to transform input IDs into dictionaries and generate_sparse_vectors to handle the tokenization and dictionary creation.
def build_dict(input_batch):
# store a batch of sparse embeddings
sparse_emb = []
# iterate through input batch
for token_ids in input_batch:
indices = []
values = []
# convert the input_ids list to a dictionary of key to frequency values
d = dict(Counter(token_ids))
for idx in d:
indices.append(idx)
values.append(d[idx])
sparse_emb.append({'indices': indices, 'values': values})
# return sparse_emb list
return sparse_emb
def generate_sparse_vectors(context_batch):
# create batch of input_ids
inputs = tokenizer(
context_batch, padding=True,
truncation=True,
max_length=512, special_tokens=False
)['input_ids']
# create sparse dictionaries
sparse_embeds = build_dict(inputs)
return sparse_embeds🚨 Note that the generate_sparse_vectors method for creating sparse vectors is not optimal. We recommend using either BM25 or SPLADE sparse vectors.
We also remove special tokens 101, 102, 103, and 0 by specifying special_tokens=False in the generate_sparse_vectors function. These are all tokens explicitly required by the BERT transformer model but have no meaning when building our sparse vectors.
This code is all we need to build our sparse vectors, but as usual, we still need to create dense vectors.
Dense Vectors
Our dense vectors are comparatively simple to generate. We initialize a multi-qa-MiniLM-L6-cos-v1 sentence transformer model and encode the same context as before like so:
# !pip install sentence-transformers
from sentence_transformers import SentenceTransformer
# load a sentence transformer model from huggingface
model = SentenceTransformer(
'multi-qa-MiniLM-L6-cos-v1'
)
emb = model.encode(contexts[0])
emb.shape(1, 384)
The model gives us a 384 dimensional dense vector. We can move on to upserting the full dataset with both sparse and dense vectors.
Creating the Sparse-Dense Index
The process for creating and using a sparse-dense index is almost identical to creating a pure dense index, the only chance being that upserts and queries must include an additional sparse_values parameter. To initialize the sparse-dense enabled index we do:
import pinecone # !pip install pinecone-client
pinecone.init(
api_key="YOUR_API_KEY", # app.pinecone.io
environment="YOUR_ENV" # find next to api key in console
)
# choose a name for your index
index_name = "hybrid-search-intro"
# create the index
pinecone.create_index(
index_name = index_name,
dimension = 384, # dimensionality of dense model
metric = "dotproduct",
pod_type = "s1"
)To use a sparse-dense enabled index we must set pod_type to s1 or p1, and metric to use dotproduct.
With all of that ready, we can begin adding all of our data to the hybrid index like so:
from tqdm.auto import tqdm
batch_size = 32
for i in tqdm(range(0, len(contexts), batch_size)):
# find end of batch
i_end = min(i+batch_size, len(contexts))
# extract batch
context_batch = contexts[i:i_end]
# create unique IDs
ids = [str(x) for x in range(i, i_end)]
# add context passages as metadata
meta = [{'context': context} for context in context_batch]
# create dense vectors
dense_embeds = model.encode(context_batch).tolist()
# create sparse vectors
sparse_embeds = generate_sparse_vectors(context_batch)
vectors = []
# loop through the data and create dictionaries for upserts
for _id, sparse, dense, metadata in zip(
ids, sparse_embeds, dense_embeds, meta
):
vectors.append({
'id': _id,
'sparse_values': sparse,
'values': dense,
'metadata': metadata
})
# upload the documents to the new hybrid index
pinecone.upsert(vectors)
# show index description after uploading the documents
pinecone.describe_index_stats(){'namespaces': {'': {'vectorCount': 1000}},
'dimension': 384,
'indexFullness': 0,
'totalVectorCount': 1000}From describe_index_stats, we should see that 1000 records have been added. With that, we can move on to querying the new index.
Making Queries
Queries remain very similar to pure dense vector queries, with the exception being that we must include a sparse vector version of our query — alongside the typical dense vector representation.
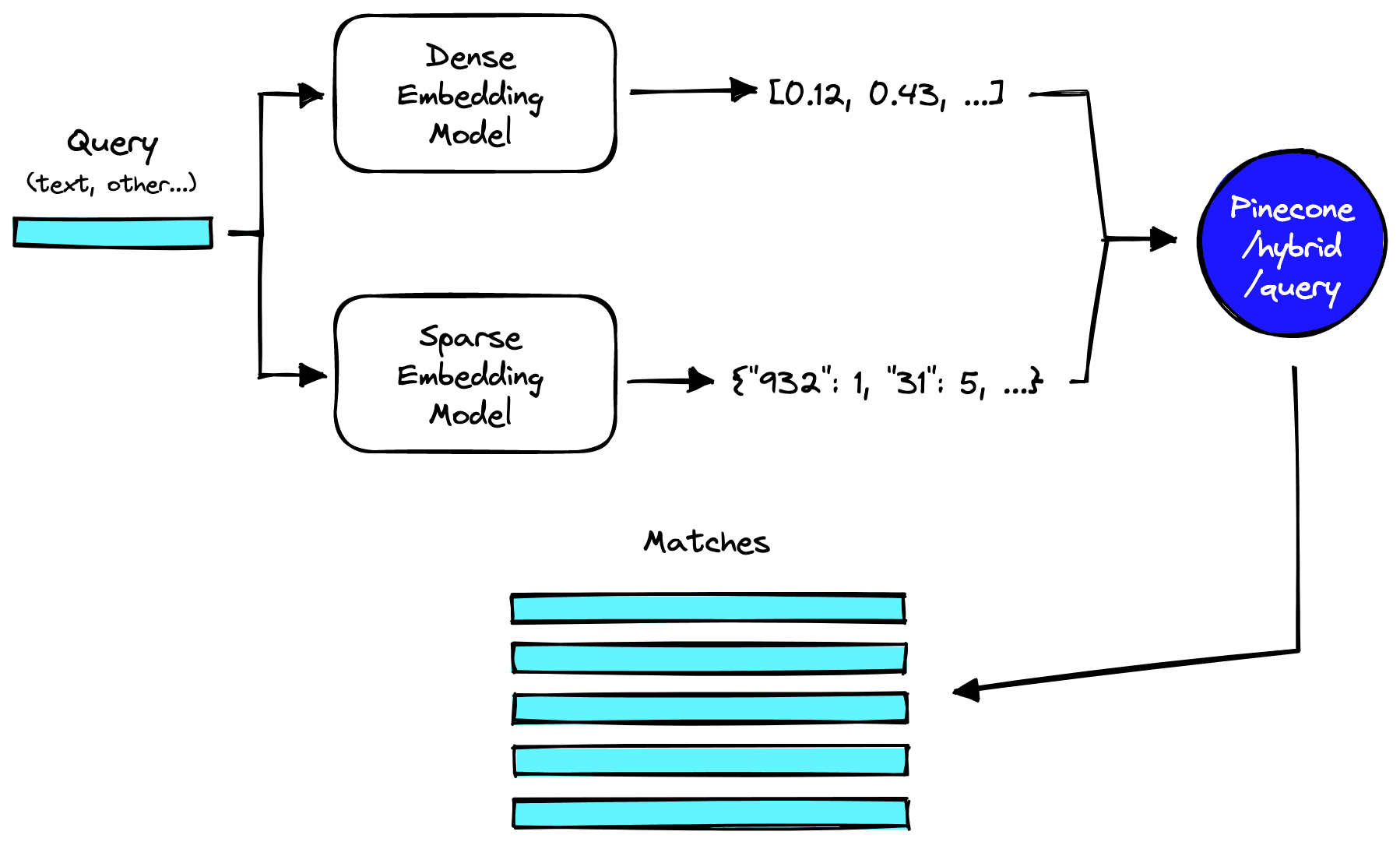
We can use the earlier generate_sparse_vectors function to build the sparse vector. We will wrap the encode and query operations into a single hybrid_query function to keep queries simple.
def hybrid_scale(dense, sparse, alpha: float):
# check alpha value is in range
if alpha < 0 or alpha > 1:
raise ValueError("Alpha must be between 0 and 1")
# scale sparse and dense vectors to create hybrid search vecs
hsparse = {
'indices': sparse['indices'],
'values': [v * (1 - alpha) for v in sparse['values']]
}
hdense = [v * alpha for v in dense]
return hdense, hsparse
def hybrid_query(question, top_k, alpha):
# convert the question into a sparse vector
sparse_vec = generate_sparse_vectors([question])[0]
# convert the question into a dense vector
dense_vec = model.encode([question]).tolist()
# scale alpha with hybrid_scale
dense_vec, sparse_vec = hybrid_scale(
dense_vec, sparse_vec, alpha
)
# query pinecone with the query parameters
result = pinecone.query(
vector=dense_vec,
sparse_vector=sparse_vec[0],
top_k=top_k,
include_metadata=True
)
# return search results as json
return resultNow we query like so:
question = "Can clinicians use the PHQ-9 to assess depression in people with vision loss?"hybrid_query(question, top_k=3, alpha=1){'matches': [{'id': '305',
'score': 0.732677519,
'values': [],
'sparseValues': {},
'metadata': {'context': 'The gap between evidence-based treatments and routine care has been well established. Findings from the Sequenced Treatments Alternatives to Relieve Depression (STAR*D)...'}},
{'id': '711',
'score': 0.696874201,
'values': [],
'sparseValues': {},
'metadata': {'context': "To investigate whether the Patient Health Questionnaire-9 (PHQ-9) possesses the essential psychometric characteristics to measure depressive symptoms in people with visual impairment..."}},
{'id': '735',
'score': 0.505706906,
'values': [],
'sparseValues': {},
'metadata': {'context': "To assess whether perspective-taking, which researchers in other fields have shown to induce empathy, improves patient satisfaction in encounters between student-clinicians and..."}}],
'namespace': ''}How can we assess the impact of hybrid search vs. vector search with these results? We use the new alpha parameter that is fed into the hybrid_scale function.
The alpha parameter controls the weighting between the dense and sparse vector search scores. By default, this is set to 0.5, making any results a pure hybrid search.
Above we performed a pure dense vector search by using an alpha of 1.
With a full vector search, we get the same ranking of results. However, the “best” context (711) is currently in position two. We can modify the alpha parameter to try and improve this result.
hybrid_query(question, top_k=3, alpha=0.3){'matches': [{'id': '711',
'score': 0.486920714,
'values': [],
'sparseValues': {},
'metadata': {'context': "To investigate whether the Patient Health Questionnaire-9 (PHQ-9) possesses the essential psychometric characteristics to measure depressive symptoms in people with visual impairment...'}},
{'id': '305',
'score': 0.461695194,
'values': [],
'sparseValues': {},
'metadata': {'context': 'The gap between evidence-based treatments and routine care has been well established. Findings from the Sequenced Treatments Alternatives to Relieve Depression (STAR*D)..'}},
{'id': '130',
'score': 0.383923233,
'values': [],
'sparseValues': {},
'metadata': {'context': "Although record linkage of routinely collected health datasets is a valuable research resource, most datasets are established for administrative purposes and not for health outcomes..."}}],
'namespace': ''}Using an alpha of 0.3 improves the results and returns the best context (711) as the top result.
Once we’re done we delete the index to save resources like so:
pinecone.delete_index(index_name)That’s it for our fast introduction to hybrid search and how we can implement it with Pinecone. With this, we can reap the benefits of dense vector search while sidestepping its out-of-domain pitfalls.
If you’d like to get started with hybrid search, it is available via private preview with Pinecone. Get in touch for early access!